Breakthrough in Protein Interaction Research: Hungarian Researchers' Findings Could Help Treat Oncological Diseases
Researchers at the Drug Innovation Centre of the HUN-REN Research Centre for Natural Sciences and the HUN-REN–SZTE Biomimetic Systems Research Group have developed a novel solution to repair protein networks damaged by diseases. Their groundbreaking findings were featured on the cover of Angewandte Chemie, one of the world’s most prestigious chemistry journals.
It is a fact that proteins perform many functions in our bodies, yet deciphering the unique and intricate workings of individual proteins remains a formidable challenge. For those who have followed the achievements and careers recognised by the 2024 Nobel Prizes, it will come as no surprise that protein research holds a central place in contemporary scientific discourse. This year’s Nobel Prize in Chemistry was awarded for groundbreaking advances in this field: Demis Hassabis and John Jumper were honoured for predicting protein structures, while David Baker received the prestigious award for designing new proteins.
Complex task, intricate answer
Even within a single microscopic cell, tens of thousands of uniquely structured proteins form networks through protein-protein interactions. These networks function much like social networks, transmitting 'messages' that influence key events within the cell. For example, these signals are involved in the creation, functioning, and eventual death of cells.
In many diseases, however, it is precisely the communication networks based on protein-protein interactions that are disrupted: connections fail to form, or unwanted interactions occur. According to the researchers, a possible remedy in these cases could involve restoring direct communication between the proteins involved, either by preventing unwanted interactions or by replacing those missing.
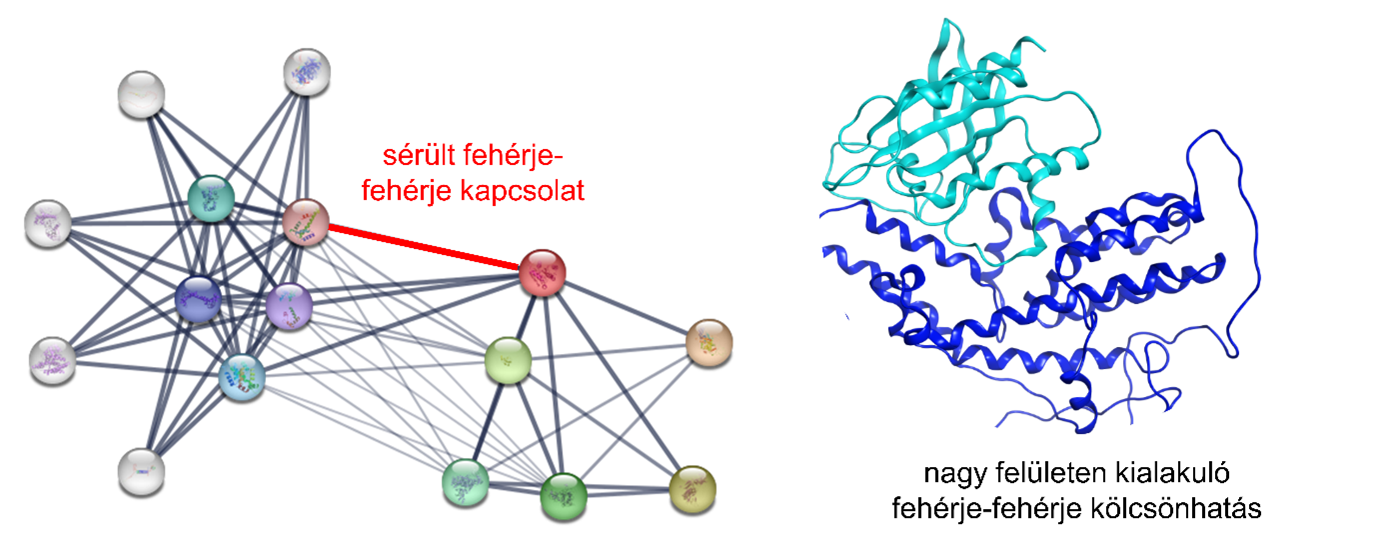
Restoring damaged connections is crucial to curing the disease. Proteomimetic molecules may be needed to prevent unwanted interactions or replace those that are missing.
However, the researchers point out that influencing protein-protein interactions is particularly challenging because these interactions not only occur over large surfaces but the interacting surfaces can also change shape. One potential solution could be to use structurally stable protein fragments derived from other proteins or interacting artificial molecules that mimic them, as they can fit onto the surface of the affected protein. Until now, however, there has been no generally applicable strategy for designing such artificial molecules.
Through the collaboration of HUN-REN working groups from three institutions, researchers have developed a universal method capable of identifying artificial molecules that mimic interacting proteins, regardless of the nature or structural characteristics of the proteins involved. To achieve this, they created a molecular library encoding the most common interaction patterns, from which the affected protein selects the interacting molecules.
How can they be identified?
The researchers used a UV light sensor to detect the binding molecules. As these molecules alter the mass of the protein, they become measurable, allowing for the mapping of interacting surfaces and the development of efficient, smaller molecules.
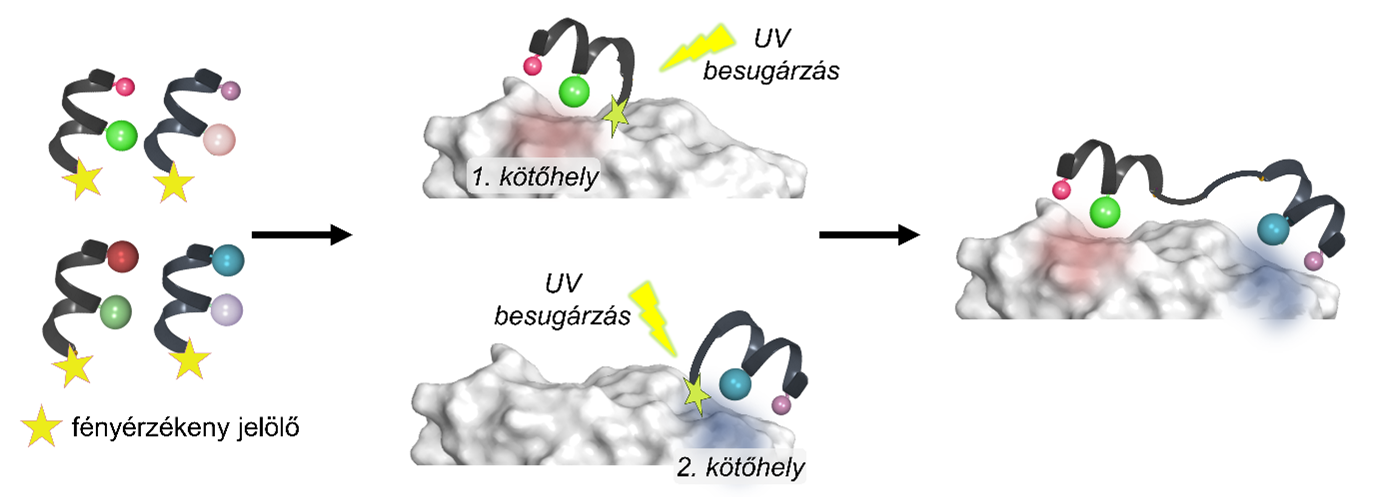
The method is based on using photosensitive markers, which are attached to the target protein by UV light, allowing the identification of their binding sites and interactions. By linking the appropriate markers, researchers can obtain molecules that mimic the effects of the proteins.
The developed method has been tested for both core applications: preventing unwanted interactions and replacing missing ones. In the context of oncological diseases, the molecules have been shown to potentially inhibit harmful protein interactions (STAT3) or replace missing partners (KRAS).
"Signal transduction pathways based on protein-protein interactions in cells play a crucial role in the development and spread of cancer. With the method we have developed, we can specifically influence these interactions and, in turn, the signalling pathways involved. In our research, we have successfully targeted proteins such as KRas, which is implicated in around 30% of tumours and can interact with multiple proteins, STAT3, which plays a role in genetic transcription, and factors responsible for the spread of certain oncogenic mutations," explains Academician György Miklós Keserű, Director of the Drug Innovation Centre at the HUN-REN Research Centre for Natural Sciences.
The research will enable researchers to characterise and validate interacting proteins – potential drug targets – that underlie diseases, even in cases where no known ligands exist. "As a result, validated drug targets will be suitable for the development of new drugs against them," says the HUN-REN researcher.
"Our results show that the developed method can identify molecules capable of repairing disease-caused defects in protein network communication by mimicking interacting partners. The strength of our method is that it can be successfully applied even in the absence of the target protein structure or without the characterisation of its interacting partners. Based on these results, we hope that it will find widespread application in drug research," summarises Tamás Martinek, Professor at the University of Szeged.

